Data Pipelines with TorchData#
In this lesson, we will generate a partitioned dataset for use with an image segmentation model. We will discuss data querying with STAC, data pipes and data loaders.
to set up the jupyter env, set up hatch as described in the readme, then:
hatch -e nb shell
exit
jupyter lab
Let’s start by importing the required packages we will use. You will notice some of them require authentication and some we use methods from without calling the package name (compositional programming).
We use this because of the following excerpts from the link above:
“If you are not careful, inheritance can lead you to a huge hierarchical structure of classes that is hard to understand and maintain. This is known as the class explosion problem.”
“Composition is more flexible than inheritance because it models a loosely coupled relationship. Changes to a component class have minimal or no effects on the composite class. Designs based on composition are more suitable to change.”
“You change behavior by providing new components that implement those behaviors instead of adding new classes to your hierarchy.”
To summarize, TorchData (a package that we will use throughout these templates) follows a compositional paradigm, where a TorchData pipe has components that change data as it moves along the pipe. Users can adapt the data pipeline by adding custom components.
# Load dependencies
import os
import urllib
import earthaccess
import geopandas as gpd
import matplotlib.pyplot as plt
import numpy as np
import pystac
import pystac_client
import shapely
import stackstac
import torch
import torchdata.dataloader2
import xarray as xr
import zen3geo
Authenticate to NASA’s Earthdata login
via earthaccess
This will prompt for a username and password the first time,
and save your credentials to a .netrc file.
auth = earthaccess.login(persist=True) # persist EDL login to netrc file
EARTHDATA_USERNAME and EARTHDATA_PASSWORD are not set in the current environment, try setting them or use a different strategy (netrc, interactive)
No .netrc found in /home/runner
Get Harmonized Landsat Sentinel-2 (HLS) raster imagery#
We will create our query to constrain a search for data hosted on NASA’s Land Processes Distributed Active Archive Center (LP DAAC).
The query consists of the following fields:
Spatial bounding box extent (bbox) in longitude/latitude coordinates
Temporal range (time_range) given by a start and end date
STAC Collection ID (collection_ids) to select the datasets in the STAC catalog
bbox = [-119.1, 36.2, -118.2, 36.9] # West, South, East, North
time_range = ["2021-08-15T00:00:00Z", "2021-09-15T23:59:59Z"]
collection_ids = ["HLSS30.v2.0"] # Harmonized Landsat-8 Sentinel-2 (HLS)
Let us setup our STAC API query to LP DAAC by putting the parameters in a Python dictionary.
query = dict(bbox=bbox, datetime=time_range, collections=collection_ids)
These parameters will be passed to PySTAC client, which will return references to the image and labels we’ll use for training later.
Reference: https://zen3geo.readthedocs.io/en/v0.6.1/stacking.html#search-for-spatiotemporal-data
dp = torchdata.datapipes.iter.IterableWrapper(iterable=[query])
dp_pystac_client = dp.search_for_pystac_item(
catalog_url="https://cmr.earthdata.nasa.gov/stac/LPCLOUD",
)
The output is a
pystac_client.ItemSearch
instance that only holds the STAC API query information but doesn’t request
for data! We’ll need to order it to return something like a
pystac.Item
using zen3geo.datapipes.PySTACAPIItemLister
(functional name list_pystac_items_by_search).
dp_hls_items = dp_pystac_client.list_pystac_items_by_search()
dp_hls_items
PySTACAPIItemListerIterDataPipe
Take a peek to see if the query does produce STAC items.
it = iter(dp_hls_items)
item = next(it)
print(item)
<Item id=HLS.S30.T11SLA.2021228T182919.v2.0>
Some of the returned HLS images may be cloudy.
We can set a filter using
torchdata.datapipes.iter.Filter
(functional name: filter)
to get only those with less than 20% cloud cover.
def cloud_cover_filter(item: pystac.Item, threshold=20) -> bool:
"""
Return True if less than or equal to 20% cloud cover, else False.
"""
return item.properties["eo:cloud_cover"] <= threshold
dp_hls_filtered = dp_hls_items.filter(filter_fn=cloud_cover_filter)
Let’s stack each of the STAC items into a 3D multispectral tensor
using zen3geo.datapipes.StackSTACStacker
(functional name stack_stac_items).
Reference: https://zen3geo.readthedocs.io/en/v0.6.1/stacking.html#stack-multi-channel-time-series-geotiffs
gdal_env = stackstac.DEFAULT_GDAL_ENV.updated(
always=dict(
GDAL_DISABLE_READDIR_ON_OPEN="EMPTY_DIR",
GDAL_HTTP_MERGE_CONSECUTIVE_RANGES="YES",
GDAL_HTTP_COOKIEFILE=os.path.expanduser("~/cookies.txt"),
GDAL_HTTP_COOKIEJAR=os.path.expanduser("~/cookies.txt"),
)
)
dp_hls_stack = dp_hls_filtered.stack_stac_items(
assets=["B04", "B03", "B02", "B08", "B12"], # RGB+NIR+SWIR bands
epsg=32611, # UTM Zone 11N
resolution=30, # Spatial resolution of 30 metres
xy_coords="center", # pixel centroid coords instead of topleft corner
dtype=np.float16, # Use a lightweight data type
rescale=False, # Don't apply scale and offset to prevent UFuncTypeError
# https://github.com/gjoseph92/stackstac/issues/133
gdal_env=gdal_env,
)
dp_hls_stack
StackSTACStackerIterDataPipe
The result is a single
xarray.DataArray
‘datacube’ with dimensions (band, y, x).
it = iter(dp_hls_stack)
dataarray = next(it)
print(dataarray)
<xarray.DataArray 'stackstac-a3d5215d42ee193cf4db55308cec210a' (time: 1,
band: 5,
y: 3784, x: 3768)>
dask.array<fetch_raster_window, shape=(1, 5, 3784, 3768), dtype=float16, chunksize=(1, 1, 1024, 1024), chunktype=numpy.ndarray>
Coordinates:
* time (time) datetime64[ns] 2021-08-19T18:54:14.997000
id (time) <U34 'HLS.S30.T11SLA.2021231T183919.v2.0'
* band (band) <U3 'B04' 'B03' 'B02' 'B08' 'B12'
* x (x) float64 2.974e+05 2.975e+05 ... 4.104e+05 4.104e+05
* y (y) float64 4.102e+06 4.102e+06 ... 3.988e+06 3.988e+06
start_datetime <U24 '2021-08-19T18:54:14.997Z'
end_datetime <U24 '2021-08-19T18:54:14.997Z'
eo:cloud_cover int64 9
title (band) <U51 'Download HLS.S30.T11SLA.2021231T183919.v2.0....
epsg int64 32611
Attributes:
spec: RasterSpec(epsg=32611, bounds=(297420, 3988410, 410460, 4101...
crs: epsg:32611
transform: | 30.00, 0.00, 297420.00|\n| 0.00,-30.00, 4101930.00|\n| 0.0...
resolution: 30
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
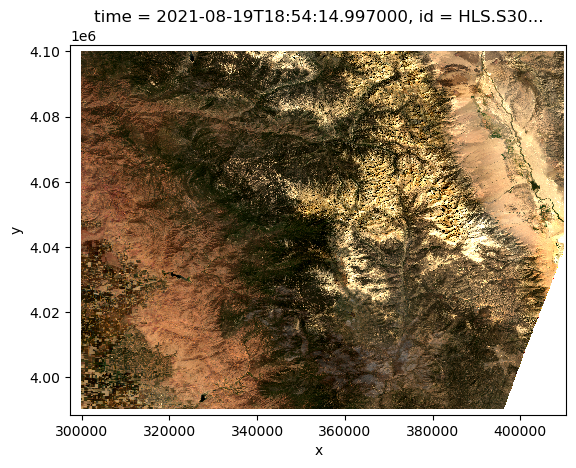
Preview a sample HLS image
dataarray.isel(time=0).sel(band=["B04", "B03", "B02"]).plot.imshow(
rgb="band", robust=True
)
<matplotlib.image.AxesImage at 0x7fe8ccbfcc70>
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
Let’s visualize the raster data pipeline so far.
torchdata.datapipes.utils.to_graph(dp=dp_hls_stack)
Get Monitoring Trends in Burn Severity (MTBS) burn scar vector labels#
Here, we will submit another query to obtain polygons of burned areas in 2021 at California. This data is available at Monitoring Trends in Burn Severity (MTBS)’s ArcGIS REST Service.
The query to the MTBS ArcGIS REST Service will be limited to the same bounding box spatial extent above.
url_base = (
"https://apps.fs.usda.gov/arcx/rest/services/EDW/EDW_MTBS_01/MapServer/77/query"
)
query = {
"geometry": "-119.1,36.2,-118.2,36.9", # xmin, ymin, xmax, ymax
"geometryType": "esriGeometryEnvelope",
"spatialRel": "esriSpatialRelIntersects",
"returnGeometry": "true",
"outFields": "*",
"outSR": "32611", # output spatial reference as EPSG:32611
"geometryPrecision": "0", # coordinates with 0 decimal places
"f": "geojson",
}
url = f"{url_base}?{urllib.parse.urlencode(query=query)}"
print(url)
https://apps.fs.usda.gov/arcx/rest/services/EDW/EDW_MTBS_01/MapServer/77/query?geometry=-119.1%2C36.2%2C-118.2%2C36.9&geometryType=esriGeometryEnvelope&spatialRel=esriSpatialRelIntersects&returnGeometry=true&outFields=%2A&outSR=32611&geometryPrecision=0&f=geojson
Accessing the URL will return a GeoJSON. However, the ArcGIS REST API service can be a bit unstable sometimes, so we’ll pull the GeoJSON from this alternative URL instead.
url = "https://gist.githubusercontent.com/weiji14/286032ac2498d10e050ba585257dd50d/raw/c897c7c1b3b8354ec8c6e8327df38fcfee79b4ef/burn_scars.geojson"
We’ll then make a HTTP request to the GeoJSON and access the byte stream using
torchdata.datapipes.iter.HttpReader
(functional name: read_from_http),
and read the polygon data via
zen3geo.datapipes.PyogrioReader
(functional name: read_from_pyogrio).
dp_url = torchdata.datapipes.iter.IterableWrapper(iterable=[url])
dp_http = dp_url.read_from_http() # outputs a tuple of (url, I/O stream)
_, dp_stream = dp_http.unzip(
sequence_length=2 # read just the I/O stream from the tuple
)
dp_pyogrio = dp_stream.read_from_pyogrio()
The polygon data will be loaded into a
geopandas.GeoDataFrame.
it = iter(dp_pyogrio)
geodataframe = next(it)
print(geodataframe)
StreamWrapper< OBJECTID ACRES FIRE_ID FIRE_NAME YEAR STARTMONTH \
0 707191 9011 CA3627811855020210815 WALKERS 2021 8
1 707192 89960 CA3658211879520210912 KNP COMPLEX 2021 9
STARTDAY FIRE_TYPE SHAPE.AREA SHAPE.LEN ... DNBR_STDDV \
0 15 Wildfire 0.003660 0.549370 ... 31
1 12 Wildfire 0.036654 2.275722 ... 39
NODATA_THRESHOLD GREENNESS_THRESHOLD LOW_THRESHOLD MODERATE_THRESHOLD \
0 -970 -150 55 283
1 -970 -150 35 313
HIGH_THRESHOLD COMMENTS LATITUDE LONGITUDE \
0 540 None 36.307 -118.546
1 610 None 36.578 -118.802
geometry
0 POLYGON ((358192.000 4022521.000, 358194.000 4...
1 POLYGON ((326156.000 4064146.000, 326115.000 4...
[2 rows x 29 columns]>
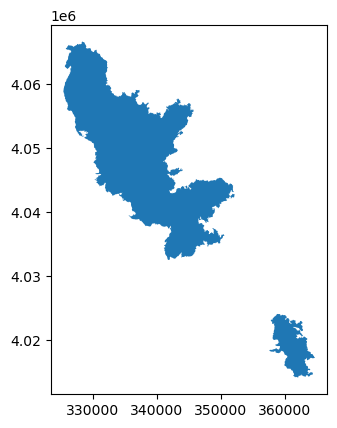
Visualize the burned area polygons.
geodataframe.plot()
<Axes: >
Data fusion of raster images and vector polygons#
In this section, we’ll work on pairing the HLS raster images with the burn scar vector polygons. This will be done in a few steps:
Create small 512x512 chips from the HLS raster images
Rasterize the burn scar polygons onto the HLS image chips
Pair image chips with their corresponding label masks
Creating 512x512 chips from large satellite scenes#
Let’s cut the large satellite scenes that are over 3000x3000 pixels in size
into smaller chips. This will be done using
zen3geo.datapipes.XbatcherSlicer
(functional name: slice_with_xbatcher)
which can slice n-dimensional datacubes along any dimensions (e.g. Eastings
and Northings).
Reference:
dp_xbatcher = dp_hls_stack.slice_with_xbatcher(
input_dims={"time": 1, "y": 512, "x": 512}
)
dp_xbatcher
XbatcherSlicerIterDataPipe
This should give us about 350 chips in total.
print(f"Number of chips: {len(dp_xbatcher)}")
Number of chips: 350
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
Rasterize vector polygons onto image chips#
Next, we’ll be painting or rasterizing the burn scar vector polygons onto each 512x512 chip, i.e. creating a binary label mask. The mask image will have the same pixel resolution and spatial extent as the chips.
Reference:
Before that, we’ll need to split our chip DataPipe into two instances using
torchdata.datapipes.iter.Forker
(functional name: fork),
one to act as an ‘canvas’ to paint the polygons on, the other one solely by
itself.
dp_chip_canvas, dp_chip_image = dp_xbatcher.fork(num_instances=2)
dp_chip_canvas, dp_chip_image
(_ChildDataPipe, _ChildDataPipe)
Now to create the canvas by calling
zen3geo.datapipes.XarrayCanvas
(functional name: canvas_from_xarray)
and paint the vector polygons onto that canvas using
zen3geo.datapipes.DatashaderRasterizer
(functional name: rasterize_with_datashader).
dp_canvas = dp_chip_canvas.canvas_from_xarray()
dp_datashader = dp_canvas.rasterize_with_datashader(vector_datapipe=dp_pyogrio)
dp_datashader
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
DatashaderRasterizerIterDataPipe
This will turn the vector
geopandas.GeoDataFrame
into a raster
xarray.DataArray
grid, with the spatial coordinates and bounds matching exactly with the
template 512x512 HLS image chip.
Pair HLS chips with label masks#
Now that we have the 512x512 HLS image chips in dp_chip_image, and the
corresponding rasterized burn scar labels in dp_datashader, we’ll need to
join them together. This can be done using
torchdata.datapipes.iter.Zipper
(functional name: zip).
dp_zip = dp_chip_image.zip(dp_datashader)
dp_zip
ZipperIterDataPipe
This creates a DataPipe which yields tuples of (image, mask) pairs. The data pipeline graph looks like so:
torchdata.datapipes.utils.to_graph(dp=dp_zip)
Some of the 512x512 chips might not have any burn scar labels (i.e. masks are
all 0 values). Once again, we can filter those out using
torchdata.datapipes.iter.Filter
(functional name: filter), while setting the input_col parameter to
examine only the mask and not the image.
def zero_mask_filter(dataarray: xr.DataArray) -> bool:
"""
Return True if input xarray.DataArray contain non-zero values, else False.
"""
return bool(dataarray.max() > 0)
dp_hls_mask_filtered = dp_zip.filter(filter_fn=zero_mask_filter, input_col=1)
dp_hls_mask_filtered
FilterIterDataPipe
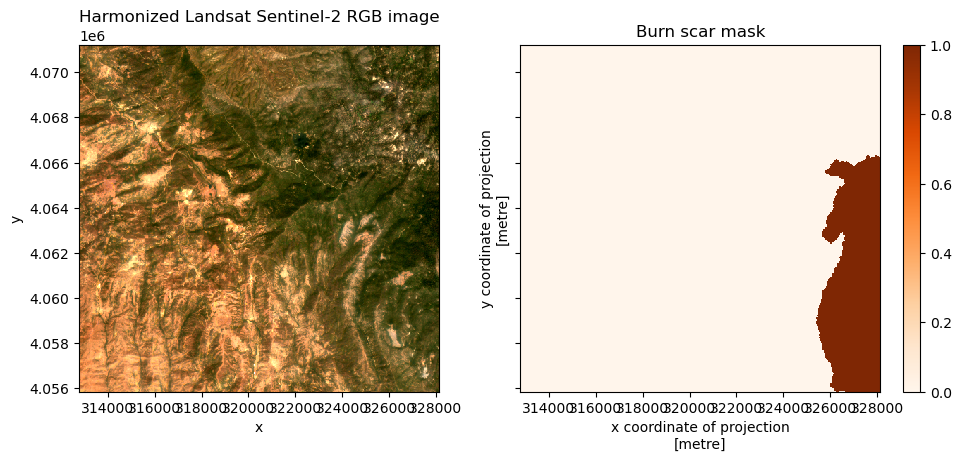
Double check to see that the HLS image and burn scar mask looks ok.
it = iter(dp_hls_mask_filtered)
hls_image, burn_mask = next(it)
# Create subplot with HLS image on the left and burn scar mask on the right
fig, axs = plt.subplots(
ncols=2, figsize=(11.5, 4.5), sharey=True, width_ratios=(1, 1.25)
)
hls_image.isel(time=0).sel(band=["B04", "B03", "B02"]).plot.imshow(
ax=axs[0], rgb="band", robust=True
)
axs[0].set_title("Harmonized Landsat Sentinel-2 RGB image")
burn_mask.plot.imshow(ax=axs[1], cmap="Oranges")
axs[1].set_title("Burn scar mask")
plt.show()
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/torch/utils/data/datapipes/iter/combining.py:297: UserWarning: Some child DataPipes are not exhausted when __iter__ is called. We are resetting the buffer and each child DataPipe will read from the start again.
warnings.warn("Some child DataPipes are not exhausted when __iter__ is called. We are resetting "
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/torch/utils/data/datapipes/iter/combining.py:297: UserWarning: Some child DataPipes are not exhausted when __iter__ is called. We are resetting the buffer and each child DataPipe will read from the start again.
warnings.warn("Some child DataPipes are not exhausted when __iter__ is called. We are resetting "
Create mini-batches to feed into a DataLoader#
Let’s finalize the data pipeline for loading into a DataLoader. From the
previous section, we have a DataPipe that yields a tuple of (image, mask)
for a single 512x512 HLS image chip and burn scar label mask. These chips
can be compiled into mini-batches using
torchdata.datapipes.iter.Batcher
(functional name: batch)
Reference:
dp_batch = dp_hls_mask_filtered.batch(batch_size=4)
dp_batch
BatcherIterDataPipe
Stack chips in mini-batch into a single tensor#
Next, we’ll need to stack all the chips within a batch into a single tensor.
A custom collate function will be used to do the conversion from many
xarray.DataArray
chips to a
torch.Tensor.
def xr_collate_fn(samples: tuple) -> torch.Tensor:
"""
Converts xarray.DataArray objects to a torch.Tensor, and stack them into a
torch.Tensor.
"""
image_tensors = [
torch.as_tensor(data=np.nan_to_num(sample[0].data).astype(dtype="int16"))
for sample in samples
]
mask_tensors = [
torch.as_tensor(data=sample[1].data.astype(dtype="uint8")) for sample in samples
]
return torch.stack(tensors=image_tensors), torch.stack(tensors=mask_tensors)
Then, pass this collate function to
torchdata.datapipes.iter.Collator (functional name: collate).
dp_collate = dp_batch.collate(collate_fn=xr_collate_fn)
dp_collate
CollatorIterDataPipe
Visualize the entire data pipeline.
torchdata.datapipes.utils.to_graph(dp=dp_collate)
Into a DataLoader#
The completed DataPipe can now be passed to
torchdata.dataloader2.DataLoader2!
dataloader = torchdata.dataloader2.DataLoader2(datapipe=dp_collate)
for i, batch in enumerate(dataloader):
image_tensor, mask_tensor = batch
print(f"Batch {i}: {image_tensor.shape}, {mask_tensor.shape}")
break
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/torch/utils/data/datapipes/iter/combining.py:297: UserWarning: Some child DataPipes are not exhausted when __iter__ is called. We are resetting the buffer and each child DataPipe will read from the start again.
warnings.warn("Some child DataPipes are not exhausted when __iter__ is called. We are resetting "
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/stackstac/prepare.py:364: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/torch/utils/data/datapipes/iter/combining.py:297: UserWarning: Some child DataPipes are not exhausted when __iter__ is called. We are resetting the buffer and each child DataPipe will read from the start again.
warnings.warn("Some child DataPipes are not exhausted when __iter__ is called. We are resetting "
/home/runner/micromamba-root/envs/mlpipeline/lib/python3.9/site-packages/torch/utils/data/datapipes/iter/combining.py:297: UserWarning: Some child DataPipes are not exhausted when __iter__ is called. We are resetting the buffer and each child DataPipe will read from the start again.
warnings.warn("Some child DataPipes are not exhausted when __iter__ is called. We are resetting "
Batch 0: torch.Size([4, 1, 5, 512, 512]), torch.Size([4, 512, 512])
The following lesson will show you how to use this data pipeline in the context of training a deep learning neural network model.


